Researchers from Wageningen have synthesised peptide-like oligomers for which the chirality of each monomer can be determined. They write in Nature Synthesis that there is huge potential for information storage.
The simplest sequence-defined chiral oligomers that we know of are peptides. They are diverse in structure and have a very specific 3D organisation that is determined to a certain degree by their chirality. By combining their shape, composition and length, they can perform specific biological functions. ‘The question for chemists becomes: can you do something similar with non-biological molecules?’ asks Han Zuilhof, Professor of Organic Chemistry at Wageningen University & Research. ‘And if so, could you expand this to create information-dense polymers?’
The group of Zuilhof and Assistant Professor Fedor Miloserdov has made significant progress in this area, as demonstrated in the Nature Synthesis paper. ‘We can even do something that nature cannot: control the configuration of the chiral centres [R or S, ed.] in the oligomer or polymer while starting with just one enantiomer of the building blocks’, continues Zuilhof. In nature, all amino acids that make up proteins are left-turning and their chirality is predefined. Miloserdov adds, ‘Our ability to determine when and how to introduce different chiralities into our oligomers is truly novel.’
Article continues below the image
64,000
The potential of these molecules is huge. ‘Consider DNA, a polymer that contains information’, says Zuilhof. ‘It has only four information-bearing units, yet DNA is one of the most densely packed information systems we know. But the molecules that our team has made could have many more units.’ Picture this: if you have four units with codons of three in a row, you’ve got 64 possible combinations. We could easily make 20 different units, which would yield a synthetic “gene” with 64,000 possibilities when using just three units!’
This idea first emerged around six years ago, but, as with most things, the path towards it was not straightforward. ‘When I joined the project, our PhD student Yu Han took on the experimental challenges’, Miloserdov recounts. ‘But there were lots of issues to overcome to achieve that ultimate goal.’
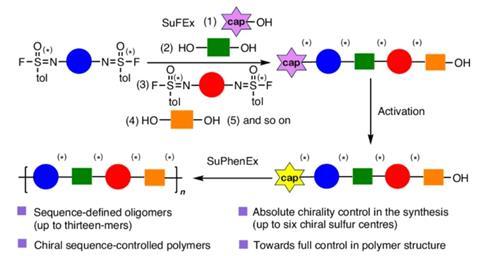
One of the problems the team faced was how to couple the molecules. ‘We wanted to do this with the SuFEx [sulphur-fluoride exchange, ed.] reaction’, Zuilhof explains. ‘It’s great for coupling one building block with another, but after a certain point, it didn’t work so well.’
As luck would have it, Zuilhof’s team had discovered a variant of the SuFEx reaction called the sulphur-phenolate exchange (SuPhenEx) reaction, completely independently from the Nature Synthesis project. When they combined both reactions, the synthesis was successful.
Hurdle
However, that wasn’t the end of the story. ‘We came across a technical hurdle that was more difficult than we expected’, says Miloserdov. When making chiral molecules, it is important to be able to prove or assess their chirality. ‘Usually, if you have one chiral centre, you can compare it to a racemic mixture. But when there is a sequence of chiral centres, the number of stereoisomers increases exponentially.’
This made things very challenging for Yu Han, who had to synthesise many other molecules as a result. ‘This was one of those PhD projects that doesn’t produce immediate results, so Yu had to be very determined’, says Zuilhof. ‘She is a remarkable woman!’
An interesting side aspect of the project was the way the chiral centres could be used to convey information. Zuilhof explains, ‘In honour of Nobel Prize laureate Barry Sharpless, we created a tetramer with the chiral centres ‘SRSS’ (ShaRpleSS). He loved it! If we had had more time, we could have made a hexamer including his first name, baRRy.’ Although it’s an inside joke, it demonstrates how information can be stored in a molecule.
Another possibility is to make sequence-controlled polymers, explains Zuilhof. ‘Many polymers simply have a pattern of aaaaaa or ababab. But we can make polymers with more complex monomer patterns such as abcd-abcd-abcd, or even one containing an e or f, which greatly enhances the potential to vary the properties.’
Now that the team has made significant progress regarding the synthesis, ‘the next big challenge is to read out the information we stored’, says Miloserdov. ‘We can read peptides because we have a mechanism to cleave them in a specific biochemical way. Something similar would be useful, and we’re already discussing ideas on how to approach this.’
Han, Y. et al. (2025) Nat. Synth., DOI: 10.1038/s44160-025-00805-8












Nog geen opmerkingen