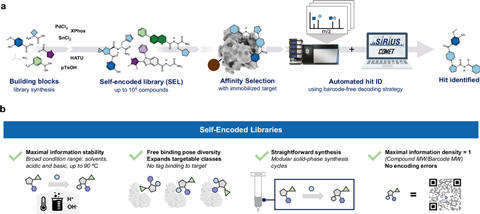
A new platform called self-encoded libraries has made the process of looking for drug candidates much simpler. This hit discovery method does not require large biomolecular tags, Leiden researchers show in Nature Comunications. ‘We believe this might democratise early drug discovery.’
The core challenge in drug discovery is looking for new molecules that could be useful in pharmacological applications. Beside classical yet resource intense industrial high throughput screenings, affinity selection platforms offer a very promising alternative. Among these, DNA-encoded libraries (DELs) stand out.
These affinity platforms use biological molecules such as DNA or peptides to label the small molecules being tested, enabling the active ones to be easily filtered out. However, these barcodes or tags may interfere with the function of the small molecule. Now, a team led by Leiden University and the Oncode Institute has developed a label-free hit discovery platform called the self-encoded library (SEL).
Femtomoles
‘During my first postdoc in medicinal chemistry and chemical biology, I attended a conference where I first became aware of the technology for identifying specific molecules within large libraries’, says Sebastian Pomplun, an associate professor at Leiden University. He coupled this idea with the way nature combats pathogens, namely by producing millions of antibodies, some of which bind to a pathogen.
Pomplun wondered if they could use chemicals alone for screening. ‘Typically, there’s a target and a large set of different molecules for screening. We check if something sticks, wash everything away and identify what remains.’ The team had to deal with screening quantities that were extremely small, sometimes as low as a few femtomoles per molecule.
‘We can identify compounds by mass spectrometry if they all have different masses. This is accessible with mass spectrometry if all the compounds are different, restricting the classical approach to small libraries’, says Pomplun. ‘However, with tandem MS, we can fragment the molecules into pieces, enabling us to distinguish many molecules that have exactly the same mass but different structures.’ In this way they can screen naked molecules in large libraries of up to half a million compounds without any barcodes, since all the necessary information is contained within the molecule itself.
Shampoo
The associate professor was very optimistic. ‘I thought we could do this on our own, so we hired a PhD student – Edith – and started the project.’ However, it turned out to be more complicated than expected, says Edith van der Nol. ‘We realised there was a lot of complexity to it. What really helped was establishing collaborations with experts in computational mass spectrometry structure annotation.’
This was a crucial step because MS can be messy. ‘It picks up everything. We can even see differences if someone used a different shampoo the night before’, says Pomplun. ‘One of the collaborators, Sebastian Boecker, saw this project as a significant opportunity. He understood the challenge and found funding to get it going.’
Decode
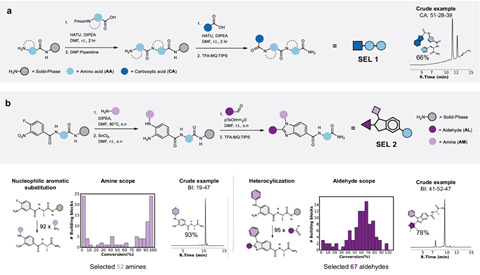
So, how does SEL work? The team used solid beads on which to build their small molecules, employing a split-and-pool synthesis method which enabled the synthesis of tens to hundreds of thousands of different compounds made up of multiple building blocks. Then, the molecules are allowed to react with a target, with some of the compounds binding to it. Tandem MS breaks the compounds down into parts that give distinct m/z values. Using a software programme, they could then decode which compound had bound to the target.
The proof of the pudding is in the eating, so Pomplun, Van der Nol and their colleagues set to work on a practical example. ‘We had extensive benchmarking to do’, says the PhD student. ‘We needed a model protein to demonstrate that our platform was functioning correctly and that we could identify the binders.’ In collaboration with the Leiden University Medical Centre, they focused on flap endonuclease 1 (FEN1). This DNA-processing enzyme would be incompatible with DNA-encoded libraries. Pomplun: ‘For the first time, we’ve been able to screen for these targets.’
Challenging
Although the method would later become successful, both Pomplun and Van der Nol were surprised by how complicated things were at the beginning. ‘Achieving clean synthesis, decoding and understanding our informatics colleagues were all challenging’, says Pomplun. ‘But when it all fell into place in a test with our biggest library, the data looked amazing and showed that it was working! This motivated us to refine the details.’
SEL sounds like a good alternative to the established protocols, but how will it find its way into the labs of those currently using DEL or other methods? Pomplun: ‘That’s exactly what we are asking ourselves. Our main focus will be improving our understanding, optimising the workflow, and getting the word out.’ At a conference in Lunteren, Van der Nol presented their research and won the presentation award. ‘There was also someone from Roche present who wanted to discuss the platform’, says Pomplun. ‘I think we’ve received more interest from industry than academia.’
The SEL platform is much easier to implement than standard techniques such as DEL or high-throughput screening. ‘It’s really simple’, says Van der Nol. ‘Every masters student who joined the project could produce hundreds, if not thousands, of compounds. This makes it interesting for industry, as we’re not competing with existing methods, but complementing them.’ Pomplun believes that ‘our platform could democratise early drug discovery.’
Van der Nol, E. et al. (2025) Nat. Commun. 16, DOI: 10.1038/s41467-025-65282-1












Nog geen opmerkingen