Under a special microscope, the spatial arrangement of DNA turns out to be more complicated than expected.
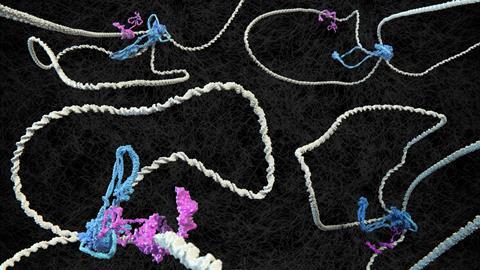
The spatial structure of chromosomes in cells is crucial for gene expression, or in other words, for reading genetic information. It is not only the genetic code itself that is important, but also the way in which the DNA is packed together at the local level. The protein cohesin is a kind of molecular motor that puts loops in the DNA. When cohesin encounters CTCF, it stops looping, or so the theory goes. But as Delft and Austrian scientists report in Nature, the real situation turns out to be more complex than expected.
Using a special microscope, the scientists managed to visualise for the first time how DNA, cohesin and CTCF interact. ‘We built it ourselves,’ says corresponding author Cees Dekker, professor at TU Delft. ‘We stuck a DNA molecule on two ends of a glass slide, gave the two proteins different colours and then we could see in the microscope whether they collided or not.’
The existing idea of CTCF as a marker that blocks cohesin was too simple. As it turned out, the effect of CTCF also depends on the DNA itself. If the DNA is under tension, CTCF stops cohesin very effectively. If the DNA is too loose, CTCF does nothing and cohesin continues to loop the DNA.
Davidson et al. (2023) Nature https://doi.org/10.1038/s41586-023-05961-5











Nog geen opmerkingen