Paracetamol is one of the very few drugs that does get completely degraded by bacteria. Nijmegen-based research now provides clues on the genes and proteins involved, and what this tells us about bacterial evolution.
A wide variety of (active ingredients of) medicines end up in wastewater each year. Because these are not natural substances, it is difficult for bacteria in wastewater treatment plants to break them down. Due to the low concentrations and variety of compounds, other removal methods are very costly. If they are available at all. As a result, pharmaceutical pollutants end up in the environment, where they accumulate. This is a problem for all kinds of organisms, including humans because these compounds eventually reappear in drinking water.
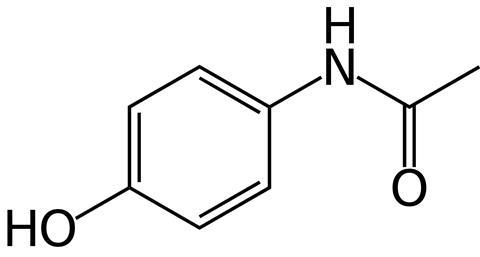
One positive exception is N-acetyl-p-aminophenol or acetaminophen, which most of us know as paracetamol. This widely used painkiller is completely degraded during the water purification process, but the responsible (bacterial) genes and proteins were still unknown. Microbiologists from Radboud University Nijmegen therefore decided to grow sludge from a hospital wastewater treatment plant on paracetamol as the only carbon source.
Amidase
By subsequently analysing this enriched bacterial community using metagenomics and metatranscriptomics techniques, they showed that several bacteria are involved in (part of the) paracetamol degradation. Cultures on paracetamol-agar plates yielded two Pseudomonas isolates. A fast-growing strain that converts paracetamol to 4-aminophenol and a slow-growing strain that completely degrades paracetamol in more than 90 days without clear intermediates. Both strains show high expression levels of an amidase, an enzyme that hydrolyses amides.
However, the researchers did not find these amidase genes in the metagenome of the sludge from the bioreactor, which they say indicates that other novel, uncharacterised amidases are also involved in the degradation process. In addition, the team saw high expression levels of deaminase and dioxygenase genes in both the bioreactor metagenome and in the two Pseudomonas genomes. Many of these genes are also part of the degradation pathways of aromatic amino acids.
Evolution
Apparently, bacteria seem to cleverly exploit the existing capacity of various enzymes to partially degrade even a non-natural substance. In addition, cross-feeding also plays a role; different species of bacteria ‘eat’ each other’s breakdown products so that a complex molecule is eventually completely broken down, while no single bacterium can do this all by itself.
The genes encoding the amidases were often located on the genome near so-called mobile genetic elements; pieces of genetic information that are easily exchanged. This suggests that horizontal gene transfer plays a role in the evolution of these bacteria to process paracetamol. According to the Nijmegen-team, these results offer new insights into how bacteria acquire the ability to break down pharmaceutical substances. At the same time, the wide variety of enzymes involved shows underlines the complexity of this process.












Nog geen opmerkingen