DNA nanotech allows you to make artificial cells that divide into different compartments without the need for membranes, British scientists show in JACS.
If you decorate artificial cells with compartments – like organelles in biological versions – you’d usually pick membranes based on polymers, fats or protein membranes. You can internally give these a heterogeneous architecture, allowing specific substances to enter or leave the ‘compartments’. However, having such an architecture in cells without any membranes was not yet available, until Adrian Leathers, Lorenzo Di Michele and colleagues at the University of Cambridge recently devised a way in which you use DNA. They managed to create up to five separate compartments and managed to get a fluorescent RNA aptamer synthesised in the nucleus that moved to the outer shell.
To build such a cell, the researchers used DNA strands. They first made a core of cross-linked DNA molecules, with a cholesterol molecule at each end. At one end there is a hook where you can attach the base strand, which makes the whole unit programmable with so-called patterned strands which differ in length. The longer the strand, the slower and stronger it will bind to the core.
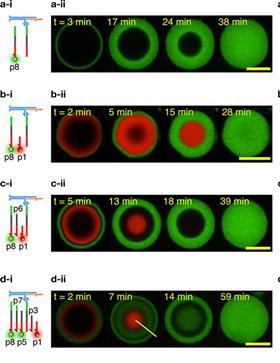
In Figure 1, for instance, you can see that the short p1 strand with a red fluorescent label reacts from the outside in. The p8 strand (green) is longer and fits onto the base strand with more base pairs than p1. Therefore, it takes longer for that strand to replace the p1 strand, but once p1 is replaced, p8 sticks much better.

If you do nothing else, the stronger binding strands will completely replace the less sticky strands, resulting in no compartments. This obviously works against the idea of compartmentalisation. There is therefore also a stopping strand: by adding a free version of the base strand in excess, you can keep the compartments in tact (see Figure 2). That’s how Leathers and colleagues managed to make a 5-compartment cell.
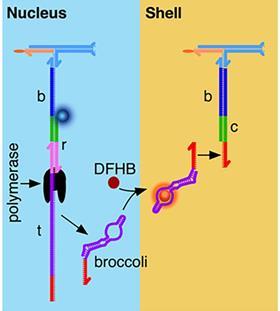
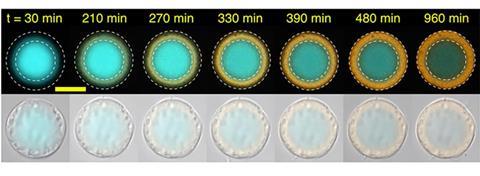
All fine and dandy, such compartments, but can you actually use them? Indeed you can, the researchers show. They made a spatially organising artificial cell with two compartments, a nucleus and a shell. They added a bridge strand and a template strand to the basic strand in the cell nucleus. The template can be read by a polymerase, resulting in a broccoli RNA aptamer. That aptamer then diffuses out of the nucleus and is captured by the base strand of the shell (see image below).
However, there were some bumps on the road. The authors indicate in their discussion that in this case you need several steps for patterning, and that you need to wash in between because the excess bridge-template complex will otherwise cause production of the aptamer in the bulk solution outside the cell. Nevertheless, with this paper, the authors think they have provided a basis for fellow scientists to build on this new platform.
Leathers, A. et al. (2022) JACS 144(38), doi.org/10.1021/jacs.2c06140












Nog geen opmerkingen