If you coat colloids with bits of specific DNA, you can use temperature to programme shapes that self-assemble. This could form the basis for new biomimetic materials, according to the authors in Nature.
For complex structures, you often need many building blocks, a bit like the different pieces of a jigsaw puzzle. That is sometimes inconvenient; preferably you would like to build complex structures with few components. New York and Paris physicists Angus McMullen, Maitane Muñoz Basagoiti, Zorana Zeravcic and Jasna Brujic looked at biology to see how proteins fold and assemble, and found a way to make complex structures with just two building blocks and some DNA strands.
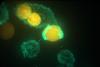
They made colloid droplets of polydimethylsiloxane in water, which they could label with fluorescent groups resulting in yellow and blue droplets. They then decorated the colloids with four different strands of DNA up to 20 base pairs long, which can interact with each other depending on temperature. Between 45 and 75°C, the droplets form a chain (backbone) of alternating blue and yellow colloids. Between 40 and 45°C activate the strong blue-blue or yellow-yellow interactions, between 30 and 35°C the slightly weaker blue-yellow interactions and around 27°C the weakest interactions between the same colours.

By playing with those DNA strands and temperature - and doing simulations on the computer - you can predict what the final outcome of the interactions will be. Thus, they calculated a whole ‘tree’ of conformations that a heptamer of four blue and three yellow colloids goes through until it eventually ends up in the last conformation if you turn on only blue-blue interactions. They superimposed the experimental plates over those of the theoretically calculated ones, and they matched.
The researchers then took their theoretical calculations one step further: simulations show that, after the initial assemblies, the structures formed can further assemble into supracolloidal systems. You then get things like mosaics, islands or - in the case of three building blocks - unique dimers.
These folding methods are very reminiscent of how proteins and peptides form, such as the hydrophobic collapse of proteins, fibril polymerisation, the formation of protein-based micelles or protein dimerisation. ‘This new paradigm of hierarchical folding as a precursor to large-scale self-assembly provides design rules for biomimetic materials with adaptable functionalities,’ the researchers conclude.
McMullen, A. et al. (2022) Nature, doi.org/10.1038/s41586-022-05198-8











Nog geen opmerkingen