Using LC-MS, extremely small protein samples enable the identification of the biological source of ivory in archeological and museum objects, Science Advances reports. ’In spite the material’s age there are still sufficient proteins present to allow a positive taxonomical identification.’
Practically since the dawn of time, cream-coloured and delicately lustrous ivory has been a particularly sought-after source material for prestige objects. Strictly speaking, the term refers to material derived from tusks, but it is also used for carved hippo or sperm whale teeth. It is often unknown what type of ivory an object is made of, or even whether it is ivory or bone. Techniques to identify the material vary in their scope and reliability. DNA analysis generally gives the most complete and reliable results, but it requires a relatively large amount of sample material and degenerates relatively quickly. As a result, this method is of limited use for old and fragile museum objects.

Proteomics may provide a solution in this case. Dental and bone material is largely composed of proteins, particularly collagen type I, encased in a mineral matrix, which helps to preserve the proteins for a longer time than DNA. The question is how small can the samples be to leave no significant trace on objects and still give a reliable result? Researchers at the University of Bordeaux, in the lab of Caroline Tokarski and the Bordeaux Proteome Platform, tested this in collaboration with The Metropolitan Museum of Art in New York and published the results in Science Advances. They sampled sixteen objects in the museum’s collection, ranging in age from several hundred to several thousand years, using diamond microgrit polishing films.
Nanograms
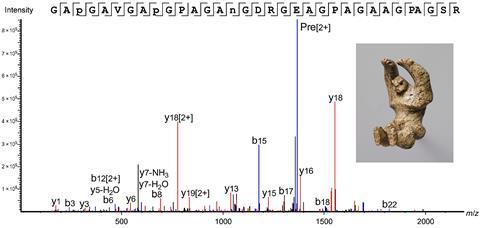
The team used LC-MS, liquid chromatography-mass spectrometry, to separate and detect peptides from the samples. They identified peptides as well as the precise amino acid sequence of those peptides, and thus the different peptides that make up the protein samples. This is the key to identification: different families, genera and species can differ by as little as a single amino acid in collagen proteins. Even in the negligibly small samples taken - effectively immeasurable at no more than a few nanograms in size - some seventy percent of the protein sequences could be retrieved with two to three rubbings at the same spot.
‘The sampling and processing show great innovation, requiring so little material’
Sebastiaan Van Nuffel
Sebastiaan Van Nuffel, assistant professor at Maastricht University and a specialist in imaging mass spectrometry, is also involved in cultural heritage research and highly impressed with the results: ‘The sampling and processing shows great innovation, requiring so little material. I was positively surprised that despite the age of the material there are still enough proteins present, and that they can lead to a positive taxonomic identification.’ Based on their analyses, the researchers were able to attribute nine of the 16 objects to a taxonomic group.
For example, keys of a 1622 virginal made of bone were found to be Bos, a genus of wild and domestic cattle, while a thirteenth-century Scandinavian ivory chess piece and an eighteenth- or nineteenth-century Hawaiian pendant could be identified as Physeter macrocephalus (sperm whale). A ceremonial saddle from the early fifteenth century was composed of several materials: bone from the pelvis of an animal of the Bos genus for the body of the saddle, while two samples from decorative trimmings were found to belong to the deer family (Cervidae).
’The amino acids leucine and iscoleucine threw everything off the rails’
Julie Arslanoglu
In some cases, it was impossible to make a more specific determination: many species simply share too many identical amino acid sequences, or publicly available databases contain sequences for no more than a handful of species. For example, the National Center for Biotechnology Information database contains sequences for only six of the total 251 members of the Cervidae family.

Elephant vs. hippo
Sometimes databases even proved completely inadequate. Ivory from ancient Egypt is typically elephant (Loxodonta africana) tusk or hippopotamus (Hippopotamus amphibius) teeth. However, reliable reference material to make a proper distinction was lacking. The researchers therefore decided to build their own proteomic database for these species with samples taken from the collections of the American Museum of Natural History in New York.

L. Africana and H. amphibius had additional challenges in store. Julie Arslanoglu, Research Scientist at the Department of Scientific Research of The Metropolitan Museum of Art and co-supervisor of the project, explains: ‘In several places, the amino acids leucine and isoleucine appeared to make the crucial difference between the two species. That threw everything off the rails: they have different chemical structures, but the same molecular mass. Was our reference material accurate or not? We had to perform an additional fragmentation step with mass spectrometry. Both amino acids lose a side chain because of that extra step, but the characteristic mass of those side chains is different and measurable. It is a tiny shift, but an absolute one.’
Contamination
An Egyptian figurine depicting an Asian captive (ca. 1295-1070 BC) also provided a surprise: after visual inspection, it had been labeled as being ivory, and the expectation was that analysis would point to hippopotamus. But the sample turned out to be collagen of the Bos genus, based on 67 characteristic peptides that were identified. However, conservators may have treated a fracture in the figurine with animal glue in the past, thus contaminating the sample. A distinction between ancient and modern collagen can be made based on the amount and kind of chemical modifications.
According to Van Nuffel, the research opens new doors in heritage research: ‘the identification of proteins lagged a bit behind until now. It now finally looks like we can start researching this properly and this bodes well for our own research into protein identification in paint layers.’ Arslanoglu is equally enthusiastic: ‘There is this whole area that is now available to us to investigate that we really couldn’t do before. We need interesting questions where the taxonomic distinction is important, where it gives you the added information about the objects you’re looking for. The method could really benefit others: archaeologists, zoologists, natural history museums. This research stimulates all kinds of new collaborations.’













Nog geen opmerkingen