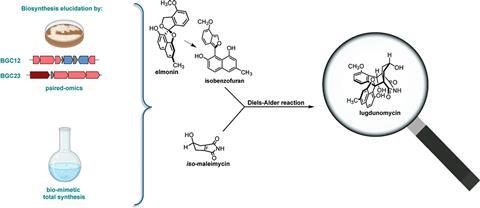
A close collaboration between teams from the universities of Leiden and Groningen succeeded in the total synthesis of the very complex molecule lugdunomycin and clarified its biosynthetic pathway, which included an intermolecular Diels-Alder reaction. They published their results in JACS.
Most drugs that enter clinical use are produced by microbes, 80% of which are based on natural products. Examples include penicillin, vancomycin and rapamycin. ‘Although we can’t seem to discover new antibiotics, there is still a lot of unexplored chemical space’, says Professor of Molecular Biotechnology, Gilles van Wezel. ‘Our labs are focused on discovering what new medicines are hiding in that space.’
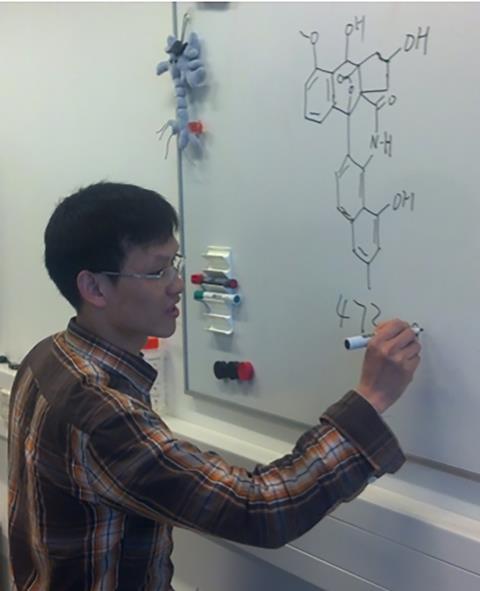
Over ten years ago, PhD student Changsheng Wu discovered a new, highly complex compound in the Streptomyces strain QL37, called lugdunomycin (named after the Latin name for Leiden, Lugdunum). ‘Despite its complexity, Wu drew the structure almost completely correctly on the whiteboard’, Van Wezel recalls. ‘But even with a thousand agar plates, we could only produce a few hundred micrograms — nowhere near enough for a comprehensive analysis.’
Missing
At that time, the NACTAR programme from NWO-TTW began, and one of these was the Lugdunomycin project, with researchers from Leiden and Groningen in collaboration with industrial partners Pfizer, Batavia Biosciences (Leiden) and Fundacion Medina (Granada, Spain). ‘Adri [Minnaard, a professor of organic chemistry in Groningen] said they would synthesise the molecule’, says Van Wezel. ‘Our aim was to understand the genetic pathway of lugdunomycin biosynthesis.’

Wu had not only predicted the structure perfectly, but also the internal pathway. ‘But one electron pair was missing. Michiel and Isabel were tasked with solving this problem.’ Michiel Uiterweerd, a PhD candidate in Minnaard’s lab, undertook the chemical synthesis, while Isabel Núñez Santiago, a PhD candidate in Van Wezel’s group, focused on the genetics and the biosynthetic pathway.
‘The realisation came after racking my brains and drawing structure upon structure’
Michiel Uiterweerd
‘When Adri showed me the molecule, I was instantly hooked. I wanted to make it!’ says Uiterweerd. It was clear at that point that a crucial step in lugdunomycin synthesis was an intermolecular Diels–Alder reaction, in which a diene reacts with a dienophile. ‘From a biochemical perspective, we first considered maleimycin as a candidate diene.’ But from a retrosynthetic perspective, that would give a Diels-Alder product with the alcohol at the wrong position.

‘We could not chemically explain how nature would then “move” the alcohol, nor did we know how to achieve that in the lab’, Uiterweerd continues. ‘So we thought an isomer of maleimycin was involved, which we coined iso-maleimycin.’ Both dienophile candidates were made in the lab, and they subsequently discovered that iso-maleimycin was produced by the QL37 strain, making it the prime candidate.
Nightmare
With a rough idea of the dienophile, they moved on to the diene. ‘One day, Helga van der Heul, another PhD student from the Van Wezel group, found a published molecule having a structural element that is also present in lugdunomycin’, says Uiterweerd. This molecule was elmonin, a known rearranged angucyclinone from Streptomyces. ‘At this point we just did not know if the QL37 strain also produced it. After racking my brains and drawing structure upon structure, I realised that elmonin looks a lot like common isobenzofuran precursor-molecules. And those isobenzofurans are highly reactive dienes in Diels-Alder reactions!’ The resulting hypothesis was that elmonin is a masked isobenzofuran, which could act as the diene.
‘By chance, I discovered that we could produce lugdunomycin in liquid cultures’
Isabel Núñez Santiago
After successfully synthesising elmonin and determining the reaction conditions required for it to react with iso-maleimycin, the nightmare of all organic chemists occurred: ‘When we compared the NMR spectrum of our compound with that of lugdunomycin, we could see all the peaks, but they had different chemical shifts. It turned out that we had made an isomer of lugdunomycin.’ Following crystallographic analysis, they discovered that it was the C9 epimer. ‘The Diels–Alder worked as predicted, but we obtained a different diastereomer than found in nature.’
Lipases
Meanwhile, theoretical chemist Remco Havenith provided computational insight into the differences between the possible pathways. It became clear that the energy differences between the reaction barriers and individual isomers weren’t that large.
‘This led us to believe that the natural reaction must involve an external factor that steers the reaction’, Uiterweerd continued. By adding some unrelated proteins to the reaction mixture, they found that some lipases altered the stereoselectivity slightly in favour of lugdunomycin and, surprisingly, could catalyse the Diels–Alder reaction. ‘Ultimately, I was able to scale up the reaction, separate the diastereomers, and isolate the molecules. This showed that iso-maleimycin and elmonin were indeed the building blocks for lugdunomycin present in QL37.’
Turning point
Finding the origin of these building blocks was Nuñez Santiago’s challenge. She built on the work of two previous PhD candidates, Helga van der Heul and Xiansha Xiao, who had focused heavily on the genetic components that the Leiden team believed to be responsible for lugdunomycin production. ‘But when our colleagues in Groningen synthesised iso-maleimycin, we could use that compound to search for it in the metabolome as well’, explains Nuñez Santiago. ‘That was a turning point for the project.’
‘The project might not have been easy, but it was great fun!’
Gilles van Wezel
‘Initially, we tried to produce lugdunomycin in liquid cultures, but it only seemed to work on solid agar plates’, Nuñez Santiago elaborates. ‘But by chance, I discovered that it did work in liquid form when galactose was used as the main carbon source.’ While conducting proteomics research, she identified proteins that were upregulated when bacterial cultures were cultivated in galactose.
It turned out that not one but two biosynthetic gene clusters were responsible for the biosynthesis of the building blocks, and they were quite far apart in the genome. ‘It is extremely unusual for such clusters to be so distant from each other yet to be engaged in the same biosynthetic pathway!’
Exception
Lugdunomycin is a highly complex molecule that occurs in multiple species of bacterium. But what does it do? ‘We simply don’t know’, admits Van Wezel. ‘It’s only a mild antibiotic, so its true function remains a mystery.’ According to the Leiden professor, his group rarely focuses on single molecules since they are mostly focusing on finding new ways and technologies to discover antibiotics. ‘But lugdunomycin is an exception. I just want to know what it does. Why would a microbe invest so much time and effort in such a complicated synthesis?’
Everyone involved really enjoyed the collaboration and found it valuable. ‘For the students, this is a real advantage: working with industry and other disciplines and growing your network. I also think it’s more fun this way, working together.’ The resulting paper essentially consists of two different stories, says Nuñez Santiago. ‘It was valuable to identify the key things we had in common and to hold sessions about the overarching story.’
Two vs. one
How will antibiotic research benefit from this work? ‘From my point of view, it’s all about knowledge’, says Núñez Santiago. ‘It’s a good example of how we need to think outside the box and look to other disciplines for collaboration. There’s still a lot of new chemistry to be discovered.’
Van Wezel hopes the work will inspire others. ‘There are many ways to find new drugs, often involving microbiology. Here, we presented a new approach. Usually, you would take the biosynthetic gene clusters, put them in an optimised host, and force the expression of one cluster to produce a molecule. But for lugdunomycin, this approach didn’t work because you need two clusters. Since then, we’ve found multiple examples of molecules that require two clusters, so using just one hides a lot of chemistry.”
‘This also provides a lot of insight into secondary metabolism’, says Uiterweerd. ‘There’s a lot of activity in this metabolism. If you want to discover new antibiotics, you need to understand these processes.’ Van Wezel adds, ‘Lugdunomycin is an angucycline, the largest family of natural products. This project enabled us to expand our knowledge of the biosynthesis of these compounds significantly, providing lots of new chemical insights. It might not have been easy, but it was great fun!’
Uiterweerd, M.T., Nuñez Santiago, I. et al. (2025) JACS 147(16), DOI: 10.1021/jacs.5c01883












Nog geen opmerkingen