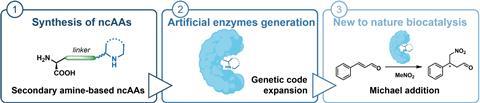
Scientists in Amsterdam have succeeded in incorporating amino acids with a secondary amine into a protein to give it an enzymatic function, reports ChemCatChem.
There are many techniques for making molecules, but one relatively ‘clean’ way is to use enzymes as catalysts for certain reactions. However, using only natural enzymes limits the range of reactions that can be carried out. Scientists have therefore been producing variants or their own versions of enzymes or proteins for years, sometimes using non-canonical amino acids (ncAAs). Alejandro Gran-Scheuch, Elisa Bonandi and Ivana Drienovská from the Vrije Universiteit Amsterdam combined the most popular organocatalytic functions with a non-enzymatic protein, creating artificial enzymes with secondary amines for the first time.
The fact that they were able to incorporate these non-canonical amino acids with secondary amines is, I think, very special,’ says Gerard Roelfes, Professor of Biomolecular Chemistry & Catalysis at the University of Groningen. You use these secondary amines in a lot of organocatalytic reactions; the beauty of this paper is that it shows that you can now catalyse these kinds of reactions with a modified protein.
In the paper, the researchers also highlight the elegant synthesis of six ncAAs. It is a fairly simple protocol with good yields, and the final steps are pretty similar for all six, with basic and Pd-catalysed hydrolysis, respectively. But how do you get enzymes to incorporate these ncAAs and work with them?
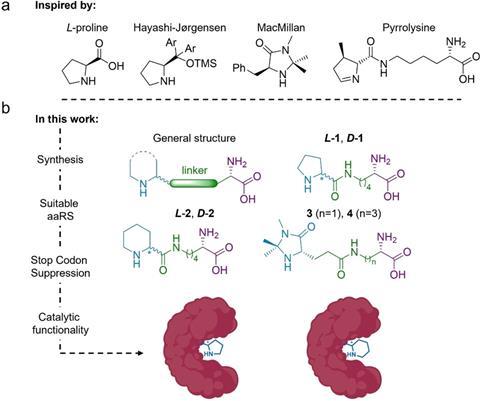
Drienovská received her PhD from Roelfes’ group, where she had previously worked with this protein (lactococcal multidrug resistance regulator, LmrR), he says. The method of inserting ncAAs into proteins has been around for about 20 years, but it has now been developed to the point where it is quite easy for chemists to use. To insert such an ncAA, a technique called stop-codon suppression is used. In the genetic material of the bacterium in which you want to produce the proteins, you reprogramme one of the three stop codons so that the ribosome reads it as a signal to insert an ncAA instead of stopping translation. To do this, you use an orthogonal translation system consisting of a tRNA from a completely different organism and an enzyme that loads the tRNA with the desired ncAA so that it is then accepted by the ribosome.
In this way, the researchers produced four different proteins; two of the ncAAs were unable to be incorporated. This gave the proteins an enzymatic function. To show that it works, the Amsterdam team carried out a reaction between nitromethane and cinnamaldehyde with and without the enzymes as catalysts. Although they only achieved reasonable yields in two variants (31 and 15%), there is no doubt that it works. This is really proof-of-principle work,” says Roelfes. You can now use directed evolution to improve this basic activity. They haven’t done that here yet, but you can scale up the activity quite a bit. So there’s an open goal, the field just needs to score.












Nog geen opmerkingen