When you build a house or a bookshelf from a warehouse, you often use predefined components. You can do the same with proteins, as American researchers show in Nature.
What if you could apply the IKEA concept to protein design? It would make building the protein structures of your choice much easier and open up many possibilities. Timothy Huddy, David Baker and colleagues at the University of Washington have shown just that in a recent Nature paper.
‘Normally, alpha helices in proteins are twisted, so two or more alpha helices combined can form structural motifs called coiled coils, similar to straps of rope’, explains Eva Bertosin, a postdoctoral researcher in Cees Dekker’s group who works on DNA origami and wasn’t involved in this study. ‘The trick is that these alpha helices are straight, which makes them work almost like bricks.’
Simplicity
The simplicity of this work is also its beauty. ‘They have built simple units based on the same element, which they can extend in any way they like’, says Bertosin. ‘There are straight blocks, angled blocks, blocks with turns or branches, all based on the same structural element. You don’t have to make the blocks from scratch each time.’ The authors show the modular construction of different structures, such as strutted double rings, protein nanocages and even nanorail track fibres.
Article continues below the image
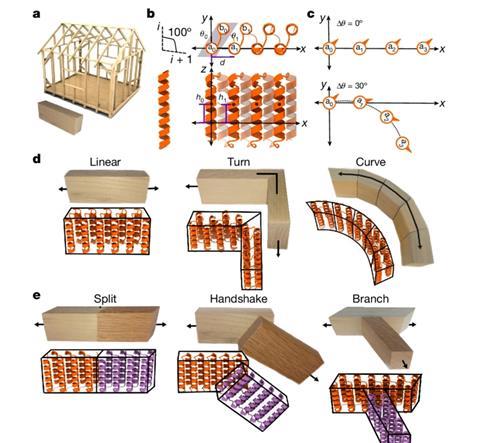
According to Bertosin, there are many ideas that could be explored. ‘You can think of protein-based materials that you make rigid by combining them with minerals. It would also be fantastic to design shapes with functionalities, such as catalytic sites or specific motions.’ Drug delivery is another possibility. ‘You could make a cage containing a drug that opens when it reaches a target cell or when it senses certain molecules.’
The ability to design structures with specific functions from scratch is a major step forward in the field. ‘If you think about natural proteins, you start with a sequence that already contains the information about its shape and function’, says Bertosin. ‘We can predict the shape, but not yet the function. But by building the function into a desired shape, as a continuation on Baker’s work may enable, you could get around that. It can also help us understand what is happening in nature and why.’
DNA origami
Bertosin uses a similar concept in her DNA origami research, where she re-engineers natural materials and tries to use them in specific contexts or give them new functionalities. ‘The field of DNA origami started a little earlier, but I think there is a possibility to combine it with de novo protein design.’
‘For example, the DNA origami field is moving towards integrated and mobile structures, and the protein design field is following that,’ explains the postdoc. ‘If you combine the motions with protein building blocks, you could make functional artificial nanomotors.’ Another possibility is to break the symmetry in Baker’s protein structures. ‘Breaking symmetry is hard, but it’s already possible in DNA origami research, so that could lead to some really interesting DNA-protein structures.’
Bertosin is really impressed and excited about the future possibilities. ‘Until a few years ago, the things that Baker and his colleagues are showing were absolutely unthinkable. And the development has been very rapid. There’s a bright future for this field, if you think about integrating functions and mimicking natural proteins, but without all the hassle that usually goes with it.’
Huddy, T.F. et al. (2024) Nature 627, DOI: 10.1038/s41586-024-07188-4













Nog geen opmerkingen