Imagine scanning a spectrum of an unknown molecule and receiving a few suggestions from an AI assistant as to what it could be. Researchers at MIT and IBM are actively working on this technology and have already achieved powerful results. ‘Those who don’t embrace AI may find themselves left behind.’
When faced with the challenge of interpreting analytical spectra, a common question echoes throughout many laboratories: ‘What in the world am I looking at?’ Fortunately, recent developments in AI, machine learning, and language models have encouraged scientists to tackle this problem using these tools. For instance, new multimodal datasets integrate different spectroscopic data, enabling machine learning models to predict molecular structures more accurately. Other examples include models that transform infrared (IR) or mass spectra into molecular structures.

Notable efforts can be found at IBM, where Teodoro Laino, a distinguished research scientist, and PhD student Marvin Alberts are working on language models. ‘We were among the first to use such models for scientific tasks around eight years ago, and we ultimately arrived at the use case of solving analytical spectra, on which Marvin works.’ Their main goal? Structure elucidation. ‘After a brief synthesis during my master’s degree, it took me over a month to carry out all the necessary measurements and characterisations’, explains Alberts. ‘So during my PhD, we’re aiming to automate this process.’
IR predictions
It all started with IR spectroscopy and predicting chemical structures from these spectra. ‘There was no guarantee that it would be possible’, says Laino. ‘But Marvin did an amazing job and recently achieved a prediction accuracy of 65 percent.’

This may not seem impressive, but achieving 65% accuracy in predicting the exact molecule you’re looking for from infrared spectra alone is pretty impressive, says Alberts. ‘Even if the structure doesn’t match due to mistakes, the predicted structure is often very close to the correct one.’
Until now, you needed relatively expensive NMR analysis to determine the structure of molecules. ‘But thanks to Marvin’s achievement, you can now use a machine that is a hundred times cheaper for this purpose’, Laino says. This could make a big difference in developing countries where NMR infrastructure is unaffordable, as a handheld infrared spectroscope coupled with AI models could be used instead.
790.000 molecules
Once it was clear that it worked for IR, the team set out to continue with NMR. However, there was far too little data to train models on. Therefore, Alberts and Laino wanted to lay the groundwork for a model that could perform automated structure elucidation. They extracted molecules from reaction data found in patents and filtered them. They then simulated the corresponding spectra for each molecule, creating a dataset of spectra for 790,000 molecules.
Article continues below the image
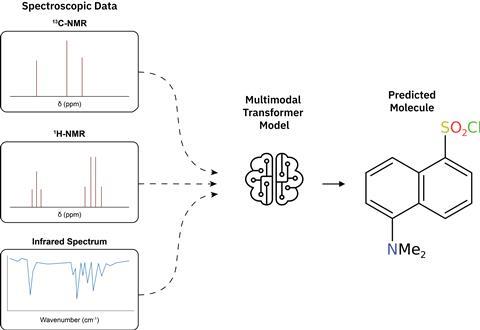
‘Based on this foundation, we began training the models’, Laino continues. ‘These models performed well, achieving results comparable to those of human chemists but much faster. They solved one spectrum in only a second.’ Using the model to provide suggestions enables chemists to validate and verify these structures much more quickly, thereby boosting efficiency.
Laino explains that the model works in a similar way to a multimodal language model. ‘Think of captions for images.’ These models leverage knowledge of context by looking at inputs from different sources. ‘The same principle applies to our model: we expose it to different types of spectra, and it uses the information to reconstruct the structure.’ Despite the relatively small amount of data they used, the team achieved an accuracy of up to 96% this way. ‘The molecules are not overly complex, but it bodes well for the future that AI models can really aid chemists in the lab’, Alberts adds.
MS predictions

Another initiative started in the group of Associate Professor Connor Coley at MIT looks at mass spectrometry (MS) data. Postdoc Runzhong Wang explains that they were inspired by the success of AlphaFold. ‘We were convinced that we could develop machine learning tools to convert mass spectrometry spectra into molecular structures in a similar way. There is a reasonable amount of MS data available, which allowed us to develop deep learning models. We reasoned that there must be some physics that can form a bridge between molecules and their spectra.’
Montgomery Bohde, an undergraduate at Texas A&M University who is also a researcher in Coley’s group through an MSRP programme, explains how their model, DiffMS, works: ‘The underlying idea is basically the same as DALL-E, ChatGPT’s image generator.’

First, you obtain your experimental spectra and determine the chemical formula. You can then put these into DiffMS, which provides a list of candidate structures that could correspond.
‘We designed the model with this generative approach on purpose, providing multiple candidate structures’, Bohde continues. ‘It’s really hard to predict the exact structure; it’s easier to provide fifteen options, one of which might be correct. That’s a useful outcome. Furthermore, multiple molecules can have similar spectra; for example, leucine and isoleucine.’
It was particularly demanding for Bohde. ‘As a computer scientist, I had no experience in chemistry or mass spectrometry. That meant I had to spend a lot of time learning about these topics from scratch.’
Reservations

Saer Samanipour, an Associate Professor at the University of Amsterdam who wasn’t involved with either the Coley or IBM group, sees many useful features in these works. ‘I think the part related to combining spectroscopic data is very good and valid. It’s going to be helpful to understand the structure through the combination of techniques — that’s the strength of these studies.’ However, he does have some reservations. ‘I believe that arriving at a structure from individual techniques will be a bit challenging.’
Samanipour is concerned that the section focusing solely on mass spectrometry data is slightly more challenging to validate. ‘The Coley group used CFMID, which doesn’t work well for many chemicals, although it’s currently the best option available. The problem with MS is that, in most cases, there isn’t enough data to solve the structure problem.’
Accuracy
‘The DiffMS paper was indeed a very challenging project’, says Wang. ‘But thanks to Montgomery, we were among the first to be able to generate structures from a spectrum with reasonable accuracy — about ten percent instead of one percent.’ Another project that was recently published on Arxiv and BioRxiv is called ICEBERG. Rather than generating a structure from a spectrum, the model is given a structure and a spectrum is generated. The Coley group is already achieving promising results with this method.
Article continues below the image
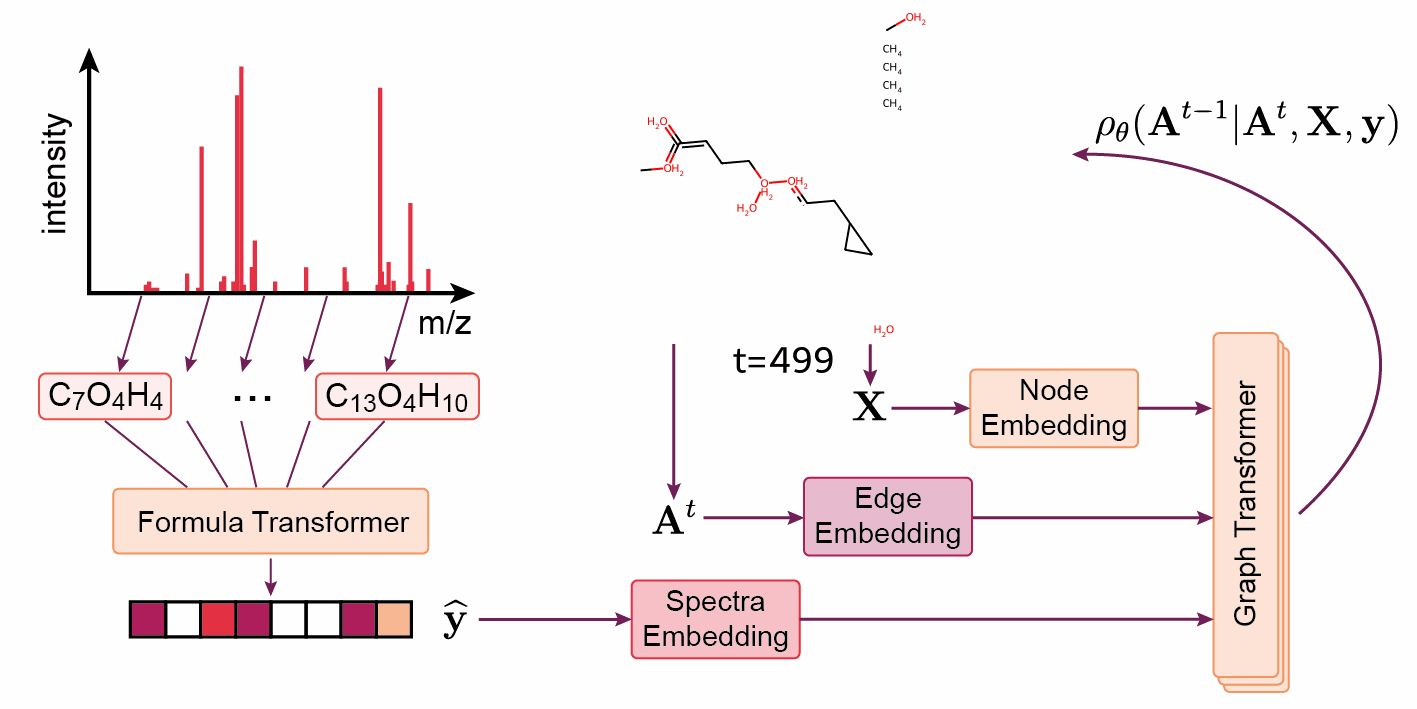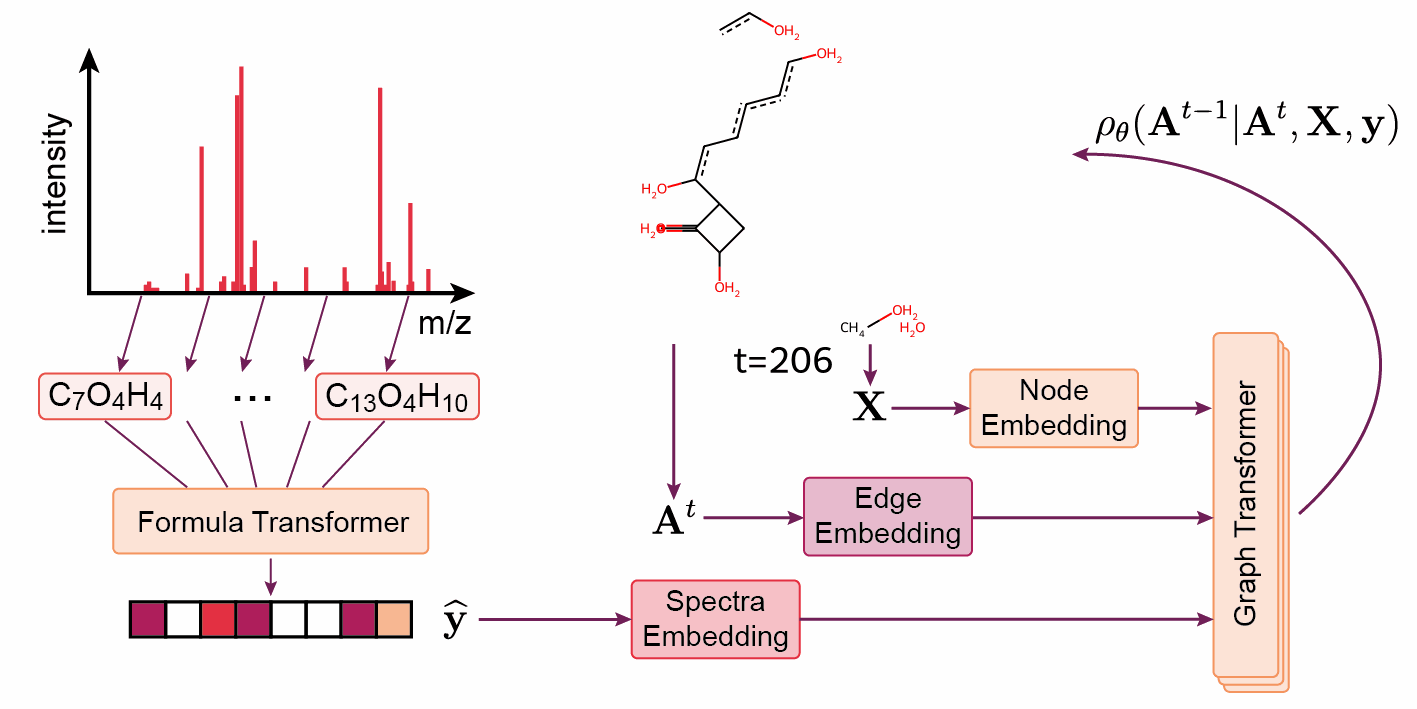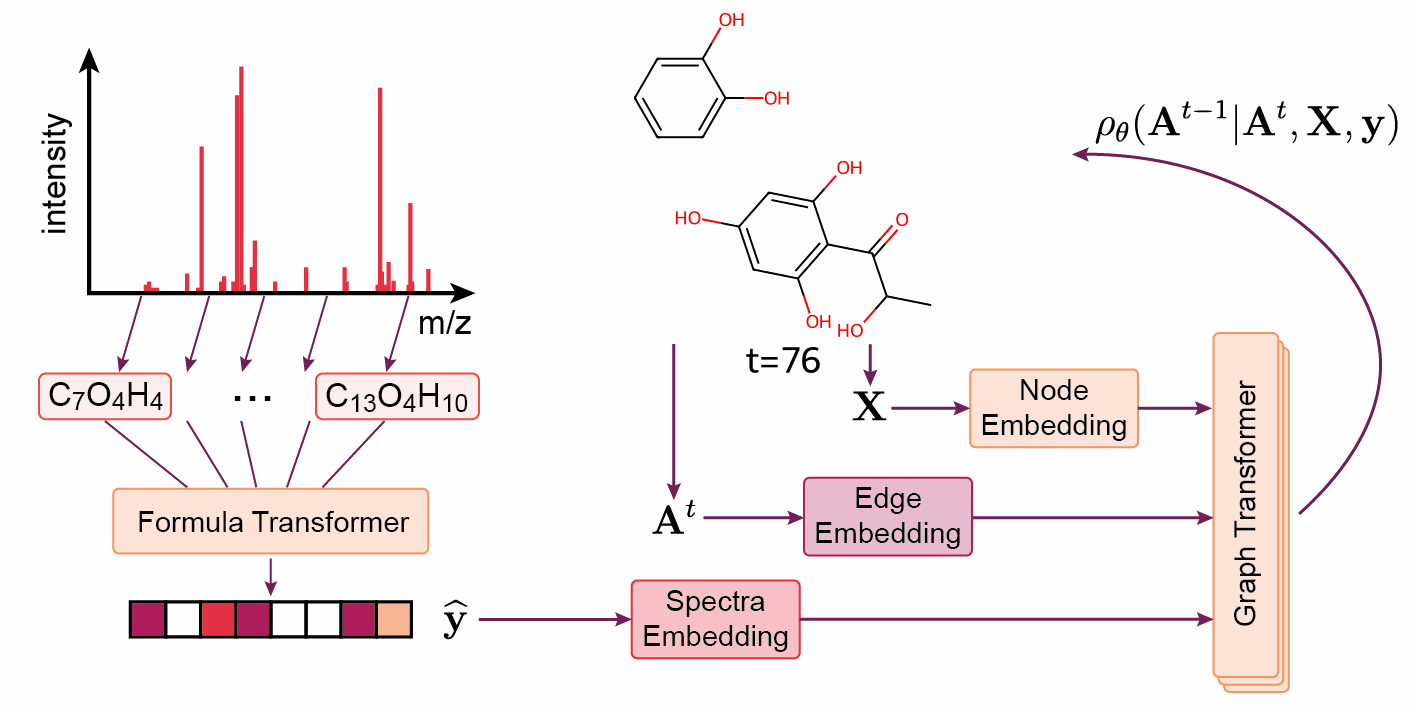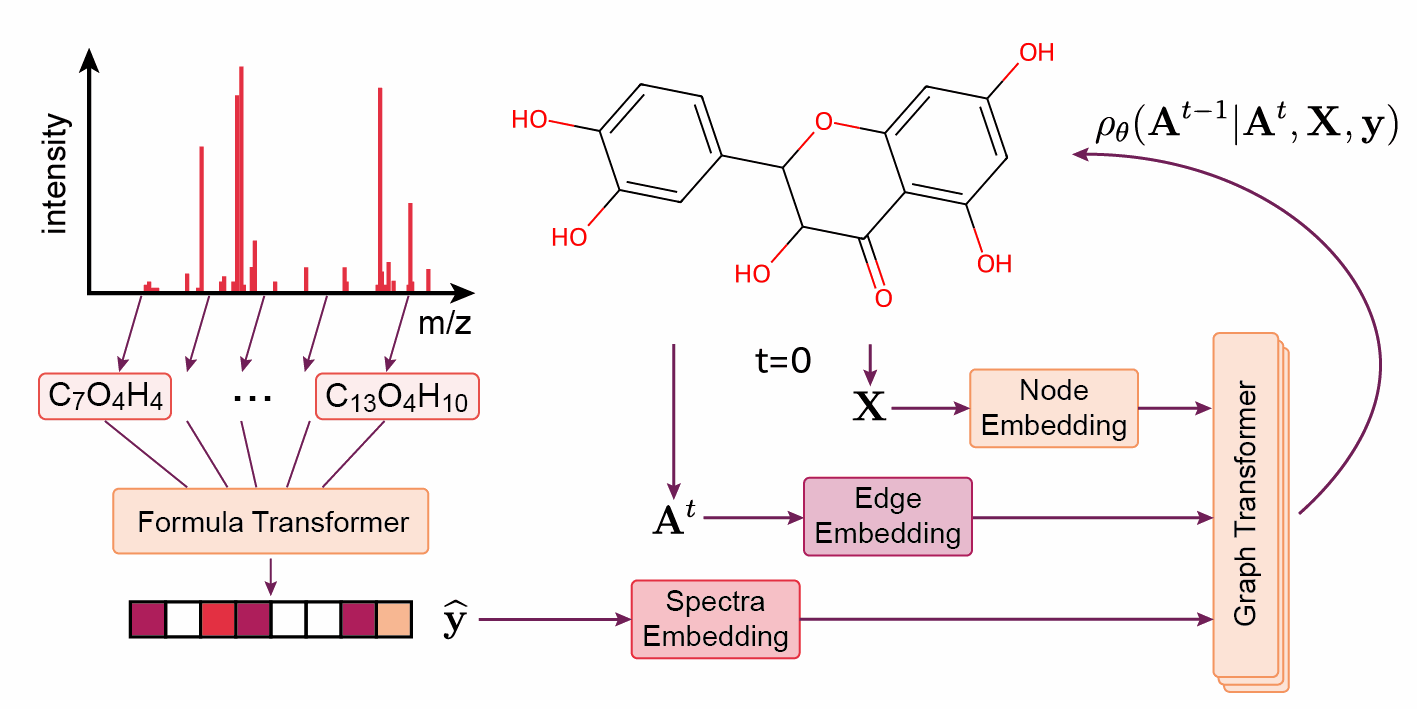
Wang continues: ‘The ideal we’re working towards is using machine learning models as another line of evidence in structure matching. Usually, you would have to buy or synthesise a ‘standard’ molecule to generate the reference spectrum, and this has to be done for every candidate molecule in every project.’ While using something like DiffMS or ICEBERG with high accuracy may not attain the same confidence level as a real standard, Wang believes it could be a viable, time- and cost-saving alternative.
Digital future
Laino believes that the future of chemistry will be digital. Chemists will have access to digital tools such as retrosynthesis design and autonomous laboratories. ‘AI won’t replace chemists, but those who don’t embrace AI and all the connected technologies may find themselves left behind.’ The discipline will evolve. How so? ‘Instead of spending time looking at spectra and trying to figure out the structure, you could just press a button and receive a few suggestions in a second.’
However, Samanipour thinks that this would mean losing valuable know-how: ‘It has always been an art to go from an NMR or MS spectrum to the structure. It’s not just about looking at the peaks; it’s also about having knowledge of the measurement environment and how the analysis was done.’ He says that there is always an issue with losing the art when you start using automatic techniques. ‘Think of arithmetic: we use our phones to calculate even simple things. The art of doing it in your head has virtually disappeared. Simultaneously, we save time or increase speed.’
‘The output should be treated as a suggestion to be assessed’
Marvin Alberts
Laino sees it differently. ‘I’m a real proponent of introducing these techniques as early as possible in education’, says the IBM researcher. You can see this in writing: it has become much easier than it was a few years ago, thanks to AI tools. ‘We should expose students to this technology while also teaching them to think critically about the process and equipping them with the skills to judge the technology.’

Alberts adds: ‘The model is still not infallible. The output should be treated as a suggestion to be assessed. I don’t believe that we will lose the ability to elucidate structures. It will evolve, but chemists will still need to understand how spectra work and how to interpret them.’
Interfering signals
A fair criticism of these works is that these technologies are used to generate molecules based on spectra of single compounds. ‘As soon as you have a spectrum containing a mixture, it becomes too complicated to decipher the molecule because interfering signals can mislead you’, says Samanipour.
However, the teams have considered this challenge. Wang: ‘It’s a good point! We assume a situation in which all compounds have been separated by both liquid chromatography and MS1 – that is different precursor mass values – so that each mass spectrum you are looking at is a single compound.’ Of course, it is possible that there are compounds co-eluting, but Wang believes that there are experimental techniques in LC-MS/MS that can potentially resolve such issues.
‘ I genuinely think that databases like this could be used to predict the fate and behaviour of chemicals’
Saer Samanipour
Alberts is optimistic about this issue: ‘We’re actually developing an approach to handle mixtures at the moment. Our work focuses on elucidating the structures of mixtures using IR spectra, and we’ve achieved surprisingly good results. We’re preparing this work for publication and have just released a preprint.’
Despite his reservations, Samanipour is convinced of the potential of these techniques. ‘From simple synthetic chemistry to drug discovery, analytical chemistry, metabolomics and exposomics, all these fields deal with identifying unknowns. I genuinely think that databases like this could be used to predict the fate and behaviour of chemicals, which would open a lot of doors to a better understanding of the exposome. It’s good to have it open and public. I hope we can take advantage of these datasets soon.’













Nog geen opmerkingen